-Search query
-Search result
Showing 1 - 50 of 215 items for (author: diamond & ms)
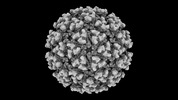
EMDB-42187: 
Eastern equine encephalitis virus (PE-6) VLP (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42189: 
Eastern equine encephalitis virus (PE-6) VLP in complex with full-length VLDLR (icosahedral)
Method: single particle / : Adams LJ, Fremont DH
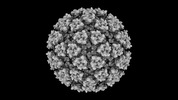
EMDB-42191: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-2) (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42193: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-3) (icosahedral)
Method: single particle / : Adams LJ, Fremont DH
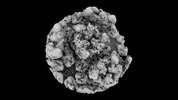
EMDB-42194: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-3) (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42195: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-5)[W132A] (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42196: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-5)[W132A] (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH
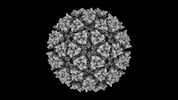
EMDB-42197: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W132A, W210A] (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42198: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W132A, W210A] (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH
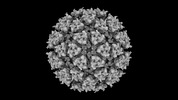
EMDB-42199: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-8)[W132A, W210A, W256A] (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42200: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-8)[W132A, W210A, W256A] (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH
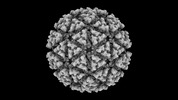
EMDB-42201: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(3-8) (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42202: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W89A] (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42203: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W89A] (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH
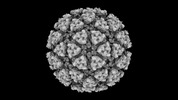
EMDB-42212: 
Eastern equine encephalitis virus (FL93-939) VLP in complex with VLDLR LA(1-6) (icosahedral)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42188: 
Eastern equine encephalitis virus (PE-6) VLP (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42190: 
Eastern equine encephalitis virus (PE-6) VLP in complex with full-length VLDLR (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH

EMDB-42192: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-2) (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH

PDB-8ufa: 
Eastern equine encephalitis virus (PE-6) VLP (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH, Center for Structural Biology of Infectious Diseases (CSBID)

PDB-8ufb: 
Eastern equine encephalitis virus (PE-6) VLP in complex with full-length VLDLR (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH, Center for Structural Biology of Infectious Diseases (CSBID)

PDB-8ufc: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-2) (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH, Center for Structural Biology of Infectious Diseases (CSBID)

EMDB-40711: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-8sqn: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID)

EMDB-27272: 
CryoEM structure of Western equine encephalitis virus VLP
Method: single particle / : Zimmerman MI, Fremont DH
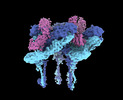
EMDB-27271: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH

EMDB-28644: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-29530: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-29531: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-40240: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-27763: 
Chikungunya VLP in complex with neutralizing Fab 506.A08 (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-27764: 
Chikungunya VLP in complex with neutralizing Fab 506.A08 (icosahedral reconstruction)
Method: single particle / : Adams LJ, Fremont DH

EMDB-27765: 
Chikungunya VLP in complex with neutralizing Fab 506.C01 (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-27766: 
Chikungunya VLP in complex with neutralizing Fab 506.C01 (icosahedral reconstruction)
Method: single particle / : Adams LJ, Fremont DH

EMDB-27767: 
Chikungunya VLP in complex with neutralizing Fab CHK-265 (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-27768: 
Chikungunya VLP in complex with neutralizing Fab CHK-265 (icosahedral reconstruction)
Method: single particle / : Adams LJ, Fremont DH

PDB-8dww: 
Chikungunya VLP in complex with neutralizing Fab 506.A08 (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-8dwx: 
Chikungunya VLP in complex with neutralizing Fab 506.C01 (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-8dwy: 
Chikungunya VLP in complex with neutralizing Fab CHK-265 (asymmetric unit)
Method: single particle / : Adams LJ, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-29020: 
Structure of dengue virus (DENV2) in complex with prM12, an anti-PrM monoclonal antibody
Method: single particle / : Dowd AD, Sirohi D, Speer S, Mukherjee S, Govero J, Aleshnick M, Larman B, Sukupolvi-Petty S, Sevvana M, Miller AS, Klose T, Zheng A, Kielian M, Kuhn RJ, Diamond MS, Pierson TC

EMDB-29021: 
Structure of dengue virus (DENV2) in complex with prM13, an anti-PrM monoclonal antibody
Method: single particle / : Dowd AD, Sirohi D, Speer S, Mukherjee S, Govero J, Aleshnick M, Larman B, Sukupolvi-Petty S, Sevvana M, Miller AS, Klose T, Zheng A, Kielian M, Kuhn RJ, Diamond MS, Pierson TC

EMDB-26200: 
Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin
Method: single particle / : Kwon YD, Gorman J, Kwong PD
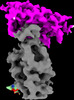
EMDB-26201: 
Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin (Local refinement of FSR22 and RBD)
Method: single particle / : Kwon YD, Gorman J, Kwong PD
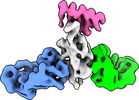
EMDB-27749: 
Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022
Method: single particle / : Kwon YD, Gorman J, Kwong PD
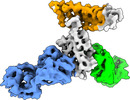
EMDB-27750: 
Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022
Method: single particle / : Kwon YD, Gorman J, Kwong PD

EMDB-25487: 
SARS-CoV-2 Spike NTD in complex with neutralizing Fab SARS2-57 (local refinement)
Method: single particle / : Adams LJ, Fremont DH

EMDB-25488: 
SARS-CoV-2 Spike in complex with neutralizing Fab SARS2-57 (three down conformation)
Method: single particle / : Adams LJ, Fremont DH

EMDB-24894: 
Ab16 Fab in complex with SARS-CoV-2 Spike (6P)
Method: single particle / : Windsor IW, Hauser BM, Schmidt AG

EMDB-24895: 
Ab20 in complex with SARS-CoV-2 spike (6P)
Method: single particle / : Windsor IW, Hauser BM, Schmidt AG

EMDB-13739: 
Tomogram of a TauRD-YFP aggregate in a TauRD-YFP expressing Hek293T cell
Method: electron tomography / : Guo Q, Saha I, Fernandez-Busnadiego R, Hartl FU

EMDB-13740: 
Tomogram of a TauRD-YFP aggregate in a TauRD-YFP expressing primary mouse neuron
Method: electron tomography / : Trinkaus VA, Saha I, Fernandez-Busnadiego R, Hartl FU
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
